research
- Precision oncology of spatial immune escape mechanisms
- Modeling of the tumor-immune microenvironment using patient-derived immune competent cultures
- Ovarian cancer specific genetic test for homologous recombination deficiency
- Integration of spatial and single-cell transcriptomic tumor-immune microenvironment
- Immunogenomic profiling to discover effective combination immunotherapies for ovarian cancer
- A new tool for cell-type annotation from single-cell multiplexed imaging and cytometry data
- Agile workflow for interactive analysis of mass cytometry data
- Single-cell spatial atlas of high-grade serous ovarian cancer
- Single-cell spatial characterization of Tertiary lymphoid structures in ovarian cancer
- Immunogenomic profiling determines responses to combination of PARP plus immunotherapy
- Our Collaborators

Precision oncology of spatial immune escape mechanisms
Surgery and chemotherapy are currently standard-of-care therapies in ovarian cancer, despite the high relapse rates and poor overall survival. Immunotherapies are being developed as an alternative or supporting therapy, but despite the promising clinical trials results, many patients fail to respond to them. The immune escape mechanisms (IEMs) are still poorly understood in ovarian cancer.
With the usage of spatial genomics, transcriptomics and single-cell immunoprofiling techniques based on tissue cyclic immunofluorescence (tCycIF), it will be possible to link ovarian cancer’s alterations with the extensive information on tumour microenvironment (TME) of the same microregion of tumour sample. This research project aims to discover the spatial IEMs within ovarian cancer tissue samples and identify the underlying genetic mechanisms. This will help to find more effective ways to target specific IEMs in ovarian cancer and to stratify patients for immunotherapies.
Modeling of the tumor-immune microenvironment using patient-derived immune competent cultures
Responses to single-agent immunotherapies have remained modest in high-grade serous ovarian cancer (HGSC), highlighting the need for combinatorial approaches. However, preserving the immune microenvironment of the native tumor under ex vivo condition remains challenging.
To address this challenge, we have established HGSC patient-
derived immunocompetent cultures (iPDCs) using a novel humanized matrix. In addition, we have optimized methods to evaluate functional and cytotoxic drug responses at single-cell level from iPDCs to combinations of DNA damaging and immunotherapeutic agents.
Overall goal of the project is to provide a platform for high-content testing of novel immuno-oncology drugs and combination treatments in HGSC.
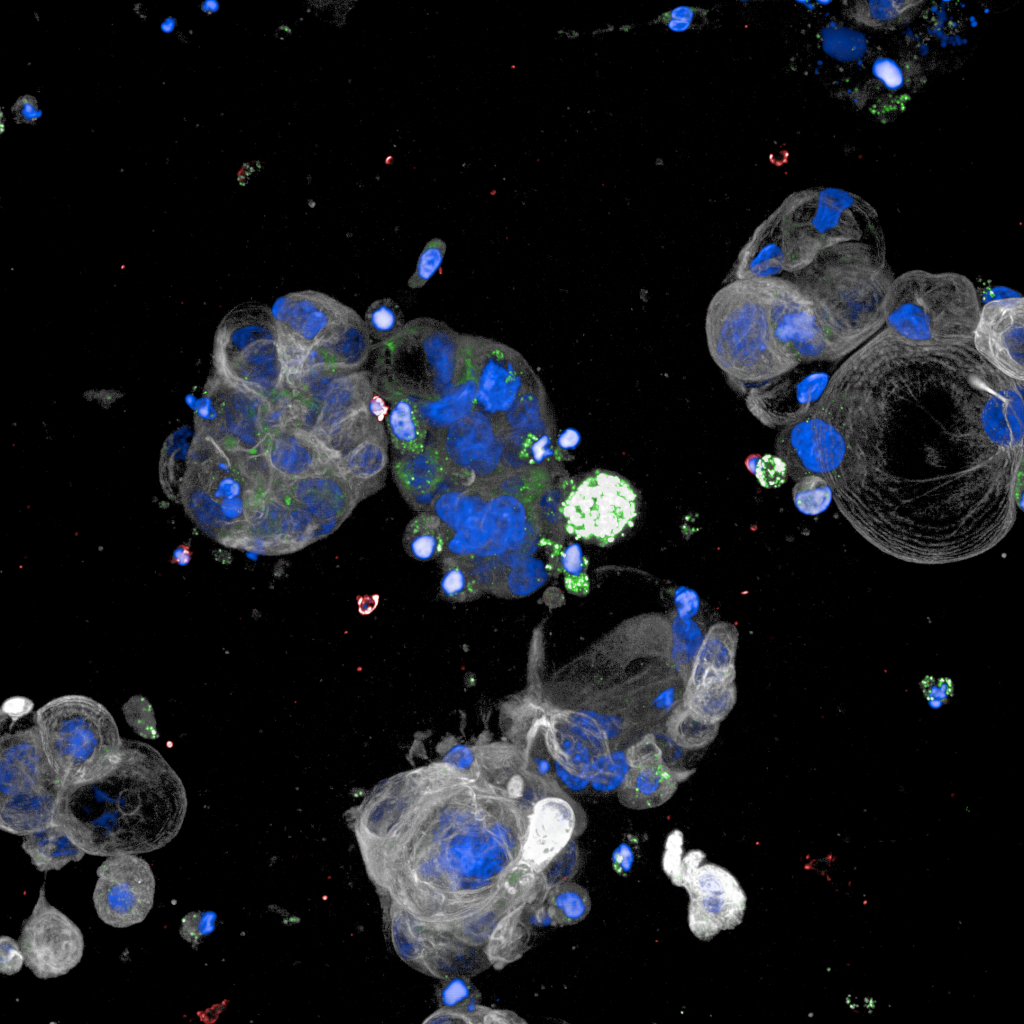
Ovarian cancer specific genetic test for homologous recombination deficiency
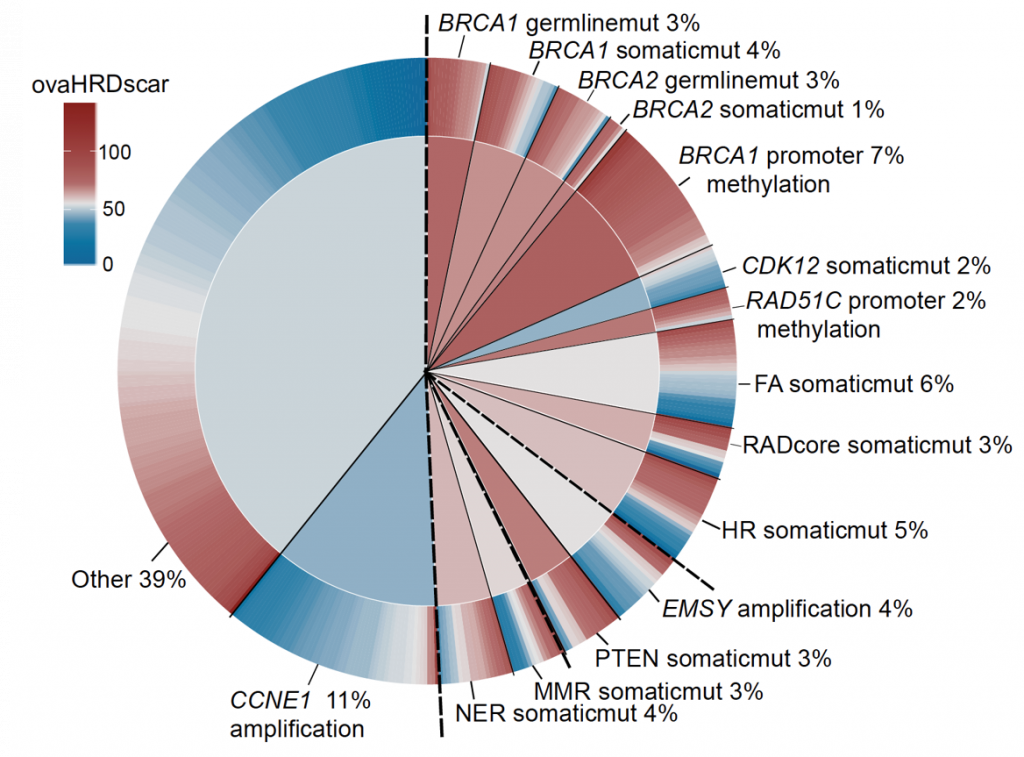
Nowadays Poly-ADP Ribose Polymerase (PARP) inhibitors have impacted the treatment of patients with ovarian cancer. During 2019 two clinical trials PRIMA and PAOLA reported that patients with tumor deficiencies of homologous recombination DNA repair (HRD), get significant benefits from PARP inhibitors as first-line maintenance therapy. Although the test used in the clinical trials was patented by a company, its cost is too high, it reports a high number of false positives, and was calibrated using a mixture of breast and ovarian cancer samples.
Using statistics and machine learning, we optimised the test for ovarian cancer (ovaHRDscar) and validated its performance in four independent cohorts.ovaHRDscar is based on the quantification of large genomic allelic imbalances specifically enriched in HRD tumours compared to proficient homologous recombination tumours. The results of the optimization are in press in the npj Precision Oncology journal, and are also available as a preprint.
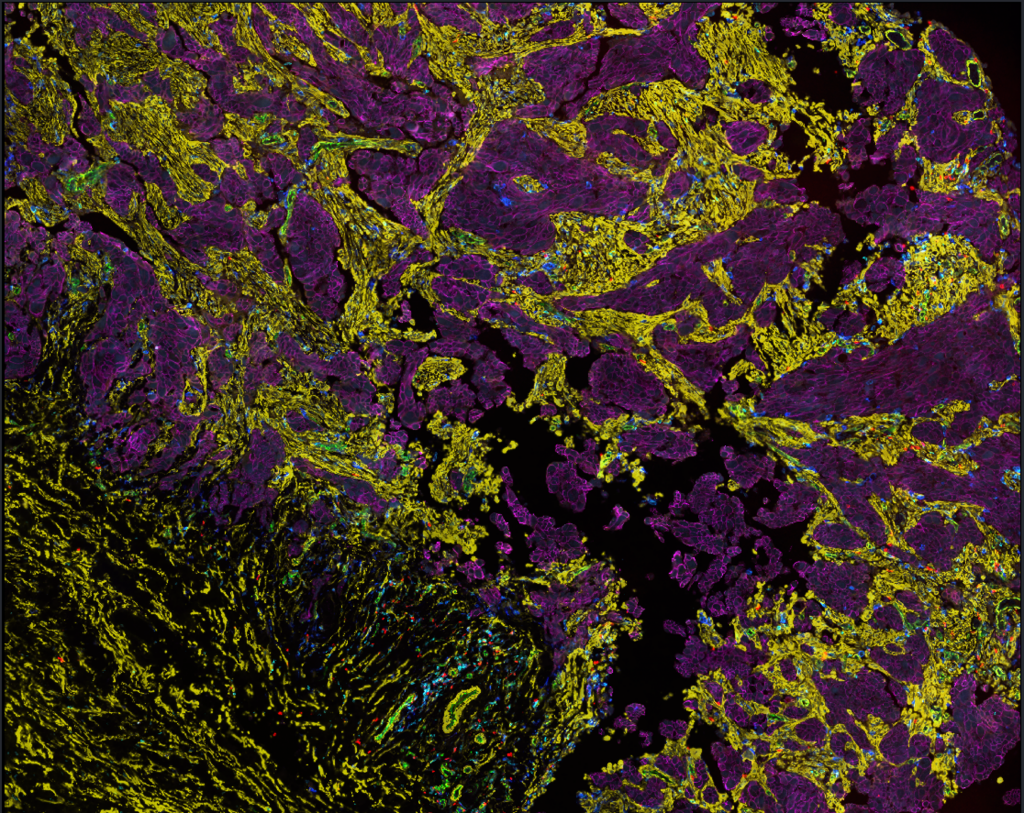
Integration of spatial and single-cell transcriptomic tumor-immune microenvironment
In the sciset project we profile the spatial dynamics of the tumor-immune microenvironment in pre- and post-chemotherapy high-grade serous ovarian cancer samples.
Using spatial statistics on t-CycIF-stained whole-slide images, we will study the effects that neoadjuvant platinum-based chemotherapy has on the immunosuppressive cell-cell interaction and avoidance patterns responsible for inferior prognosis, as well as possible immunotherapeutic targets. We will integrate these results with spatial transcriptomics, site-matched scRNAseq and whole-genome sequencing results for deep characterization of the tumor-immune microenvironment.

Immunogenomic profiling to discover effective combination immunotherapies for ovarian cancer
Oncosys-OVA is a prospective study at Helsinki University Hospital and Helsinki University, coordinated by Färkkilä Lab. We work closely with clinicians, collecting tumor tissue samples as well as clinical data of consenting ovarian cancer patients who are undergoing surgery and medical treatment in Helsinki University Women’s Hospital.
The Oncosys-OVA sample collection began in 2019, and by the end of 2022 we had close to 200 patients enrolled in the study. The collected samples and data are used in diverse projects in the lab, with the ultimate goal of developing a better understaning of ovarian cancer treatment responses, treatment resistance as well as uncovering potential novel biomarkers and treatment options for ovarian cancer patients.
A new tool for cell-type annotation from single-cell multiplexed imaging and cytometry data
Multiplexed imaging at single-cell resolution is becoming increasingly useful to decipher the role of cellular microenvironment in cancer and other complex diseases. To identify spatial patterns of single cells on a tissue we must first assign accurate descriptions to each cell in a step known as cell-type phenotyping. This step is challenging due to (i) laborious annotation of ground truth, (ii) segmentation artifacts, (iii) fluorescence noise and batch effects, and (iv) difficulty to reproduce human-biased thresholding.
Tribus is an interactive, knowledge-based classifier that avoids hard-set thresholds and manual labeling, is robust to noise, and takes less iterations from the user than standard labeling of clustering results. Interactive analysis is done via integration with the Napari image viewer, and each analysis creates a detailed report to enable reproducibility. We applied Tribus on a dataset consisting of cyclic immunofluorescence (CyCIF) images of six matched ovarian cancer samples collected before and after neoadjuvant chemotherapy. Accurate cell-type phenotyping enabled a high resolution analysis of cellular phenotypes and their spatial patterns, as well as their temporal dynamics during platinum-taxane chemotherapy. Tribus – an easily integratable open-source package – thus enables accurate phenotyping of single cells to facilitate biological discovery from highly multiplexed images.

Agile workflow for interactive analysis of mass cytometry data
We developed an agile methodology compiled as a highly comprehensive software called Cyto. Cyto makes available a curated set of data analysis tools as an optimized pipeline for single cell and cytometry analyses. This software allows rapid profiling of tumor cell subpopulations and can be used to propose actionable targets for second line therapies.
Single-cell spatial atlas of
high-grade serous ovarian cancer
Preliminary evidence suggests that DNA-repair homologous recombination (HR) deficiency tumors have a distinct tumor-immune microenvironment (TME), harboring an increased number of tumor-infiltrating lymphocytes as compared to HR-proficient tumors. In order to characterize changes in the TME by the genotypes in High-grade serous ovarian cancer, we are studying a large multi-omics dataset.
The dataset consisted of 1000 tissue microarray cores collected from both from the tumor center and from the tumor border from 250 HGSC patients. We performed cyclic immunofluorescence (tCycIF) utilizing 34 different protein markers. For each of the tumors we assessed the BRCA1/2 mutation status and the BRCA1/2 promoter hyper-methylation. For the non-BRCA1/2 mutants were performed sWGS and estimated the CCNE1 amplification status and HR-deficiency using copy number profiles and bioinformatics. The RNA expression of 340 relevant genes was assessed using Nanostring technology. The integration of multi-omics data with the single-cell spatial features will reveal the TME landscapes of HGSC with the potential to discover new biomarkers for precision oncology.

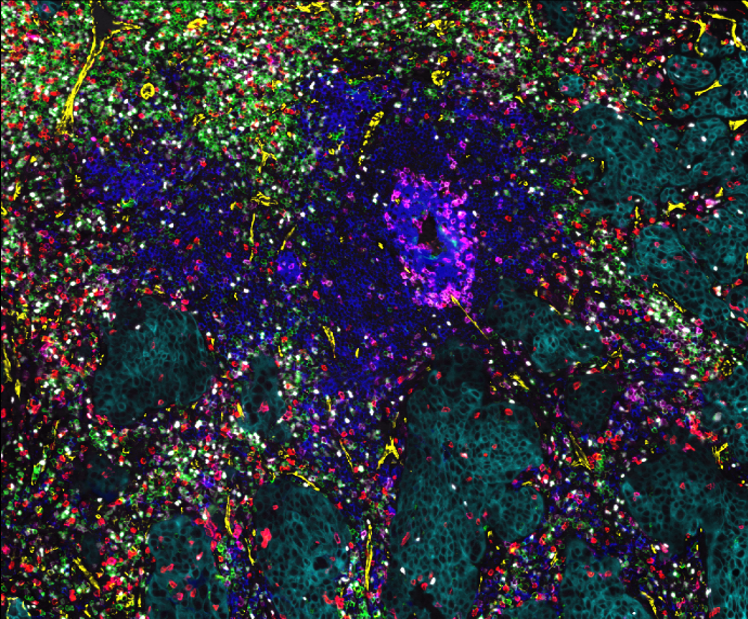
Single-cell spatial characterization of Tertiary lymphoid structures in ovarian cancer
In the TLS project, which is a collaboration project with Kekäläinen Lab, we study ectopic lymphoid aggregates called tertiary lymphoid structures (TLSs) in chemo-treated and chemo-naïve ovarian cancer patients (TOPACIO trial).
We performed more in-depth analysis of highly multiplexed immunofluorescence (CyCIF) images of ovarian tumor samples to explore cellular and spatial characteristics of TLSs. Moreover, our aim is to uncover how both treatment and homologous recombination (HR) status effect on TLSs.
Immunogenomic profiling determines responses to combination of PARP plus immunotherapy
We developed new image analysis methods to identify predictors of therapy response from tCycIF, a highly multiplexed imaging technology.
We designed spatially resolved algorithms and found that cellular proximity of PD-1 T cells and PD-L1 positive tumor cells and macrophages was highly associated with DNA damage mutational signatures
Our Collaborators






